The Science Core Facility provides necessary equipment and expertise to help maintain active research labs, and facilitate publications in peer-reviewed journals. Below are some of the most recent publications that utilized instruments in the Core.
University of Puget Sound student co-authors are in bold and faculty/post-doc/staff co-authors are italicized.
Each entry below begins with an abbreviation of the instrument(s) featured in the paper, followed by the title of the published paper.
Instrument Abbreviations
- Confocal - Nikon D-Eclipse C1 Confocal Laser Scanning Microscope
- Plate Reader - SpectraMax M2 Microplate Reader
- qPCR - Bio-Rad CFX96 Real-Time PCR Detection System
- SEM - Hitachi S3400N Variable Pressure Scanning Electron Microscope
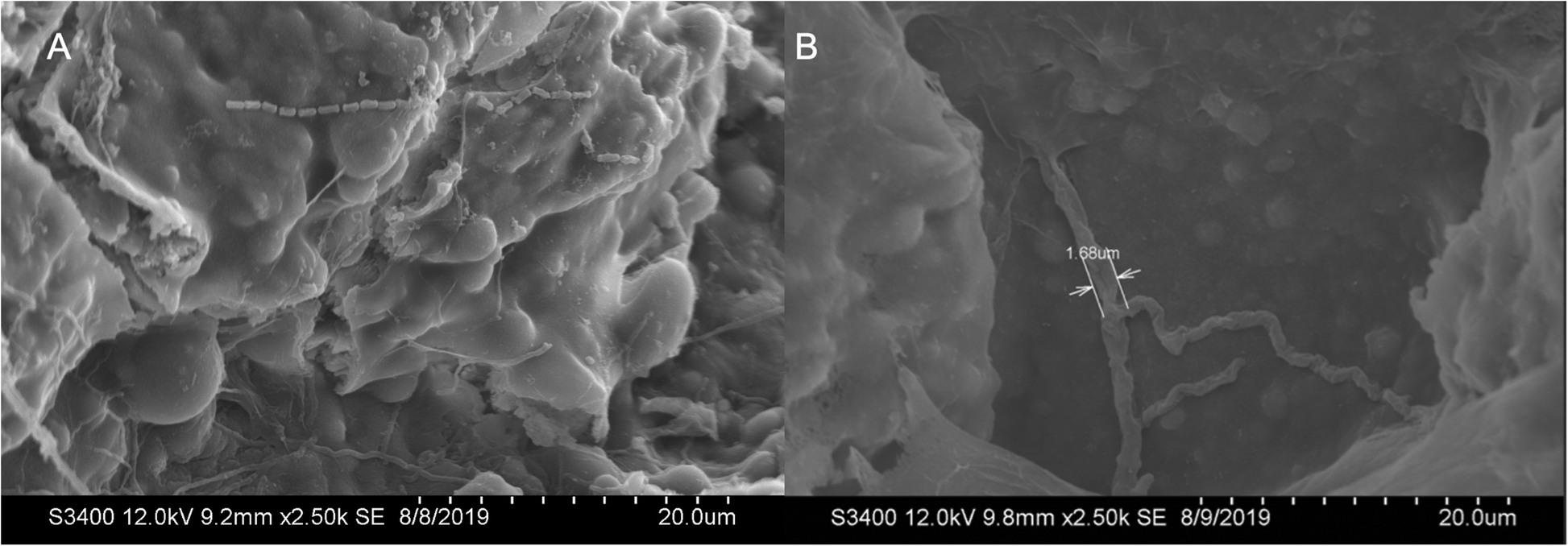
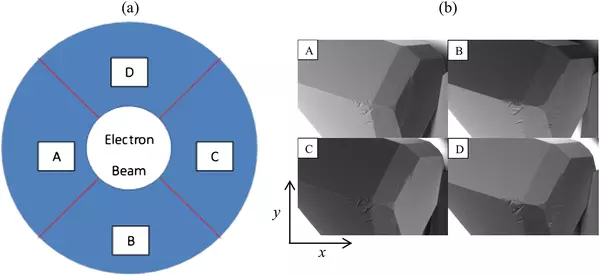
Neshyba S.P., Lowen B., Benning M., Lawson A., Rowe P.M. (2013). Roughness metrics of prismatic facets of ice. Journal of Geophysical Research: Atmospheres 118(8). https://doi.org/10.1002/jgrd.50357
- Features the use of: Hitachi S-3400N VP SEM
Pagarigan K.T., Bunn B.W., Goodchild J., Rahe T.K., Weis J.F., Saucedo L.J. (2013). Drosophila PRL-1 Is a Growth Inhibitor That Counteracts the Function of the Src Oncogene. PLOS ONE 8(4): e61084. https://doi.org/10.1371/journal.pone.0061084
- Features the use of: Nikon D-Eclipse C1 Confocal Microscope.
Matsushita S.C., Tyagi A.P., Thornton G.M., Pires J.C., Madlung A. (2012). Allopolyploidization Lays the Foundation for Evolution of Distinct Populations: Evidence From Analysis of Synthetic Arabidopsis Allohexaploids. Genetics 191(2): 535-547. https://doi.org/10.1534/genetics.112.139295
- Features the use of: Nikon D-Eclipse C1 Confocal Microscope.
Pfalzgraff W., Neshyba S., Roeselova M. (2011). Comparative Molecular Dynamics Study of Vapor-Exposed Basal, Prismatic, and Pyramidal Surfaces of Ice. Journal of Physical Chemistry 115(23): 6184-6193. https://doi.org/10.1021/jp111359a
- Features the use of: Hitachi S-3400N VP SEM
McCullough E., Wright K.M., Alvarez A., Clark C.P., Rickoll W.L., Madlung A. (2010) Photoperiod-dependent floral reversion in the natural allopolyploid Arabidopsis suecica. New Phytologist 186(1): 239-250. https://doi: 10.1111/j.1469-8137.2009.03141.x
- Features the use of: Nikon Microphot FX, Zeiss Transmission Electron Microscope EM902
Pfalzgraff W.C., Hulscher R.M., Neshyba S.P. (2010). Scanning electron microscopy and molecular dynamics of surfaces of growing and ablating hexagonal ice crystals. Atmospheric Chemistry and Physics 10: 2927-2935. https://doi.org/10.5194/acp-10-2927-2010
- Features the use of: Hitachi S-3400N VP SEM