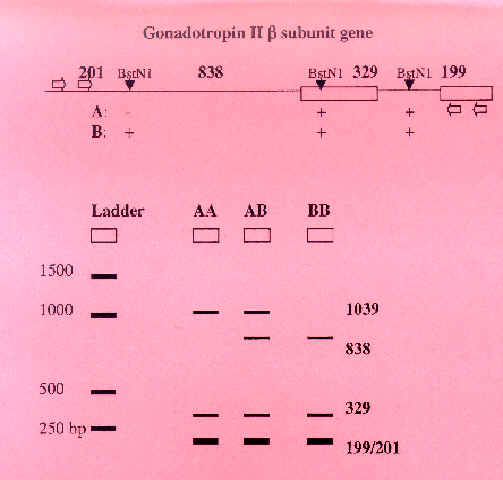
In this step, we take our PCR product (in this case, the Gonadotropin gene) and digest it with a restriction enzyme that we know cuts a site that varies among individuals. This step is one of the easiest and most satisfying if it works, because at the end of the digestion the students will have data they can analyze. We will cut GTH2B with BstN1. There are two alleles. BstN1 cuts allele A twice and allele B three times as shown in the diagram below. [A great web activity on restriction digestion (using GTH2B and BstN1 as an example) is available.]
Procedure
1. Prepare a cocktail of buffer, water and restriction enzyme for the reactions. The reactions are 20 ul and you will include 5 ul of your PCR product (from 2nd round PCR). Again, prepare enough cocktail for one extra reaction (e.g. for 9 reactions, multiply the per tube values by 10) as shown below:
Per tube e.g. x 10 (ml) (ml)ddH2O 12.5 125 10X Buffer 2 20 BstN1 0.5 5 150 (enough for 9 reactions) DNA (not in cocktail!) 5
TOTAL: 20
2. Aliquot 15 ul of the cocktail into the tube you will use for the digestion. Pipette 5 ul of your PCR product into the digest. Mix well by pipetting up and down gently, or vortexing and centrifuging.
3. Incubate your sample at 60o C for 30 - 45 minutes. (Note: BstN1 is unusual in that it works best at 60o, not 37o). You can use a thermal cycler, water bath, heat block, or incubator for the digest.
4. To visualize your samples, add 2 ul of 10X load dye directly to your digest, and run 10 ul on a 1.5% agarose gel. [Query: Why the higher percentage agarose?]
Data Analysis (analysis pages)
- Score the individual genotypes
- Calculate genotype frequencies
- Calculate allele frequencies
- Using the observed allele frequencies, calculate the genotype frequencies you would expect under Hardy-Weinberg conditions
- Use a contingency test (Chi-square) to compare the expected and observed frequencies.
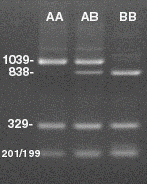
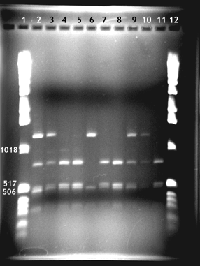
Restriction Digest Gels
(10 ul of each PCR sample digested with restriction enzyme indicated)
Gel 1: Gonadotropin Genotypes
Gel 2: Prolactin Genotypes of Rapid River Hatchery Spring Chinook Population
Lanes 1 and 12: Ladder (way too much!, and different from above)
Lanes 2-11: Prolactin PCR product cut with Hinf1
Note: The bands in the ladder under the 517/506 double band are 394,344,298, 220 bp in size. Note the faint band that is around 300 bp in length in all the digests.
Lane Genotype (prolactin)
2 AB
3 AB
4 BB
You finish! Restriction map showing size of expected fragments.